Our Process
1. Create
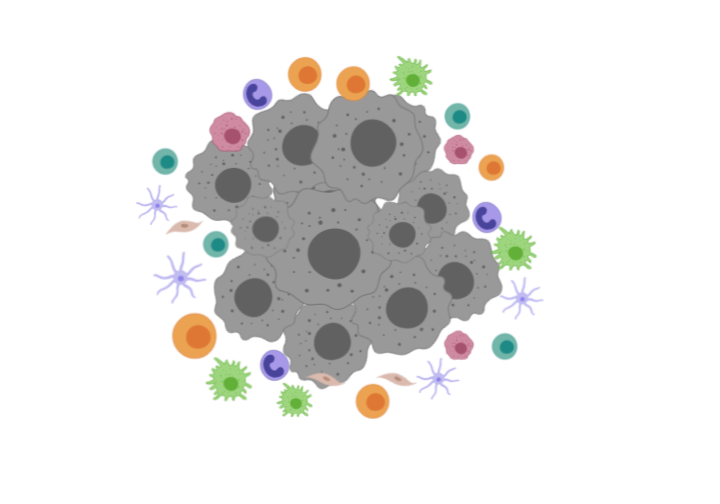
Recreate tumor environments including stromal cells, immune cells and tumors to simulate various factors that facilitate immune dysfunction and impede immune cell migration into the tumor bed.
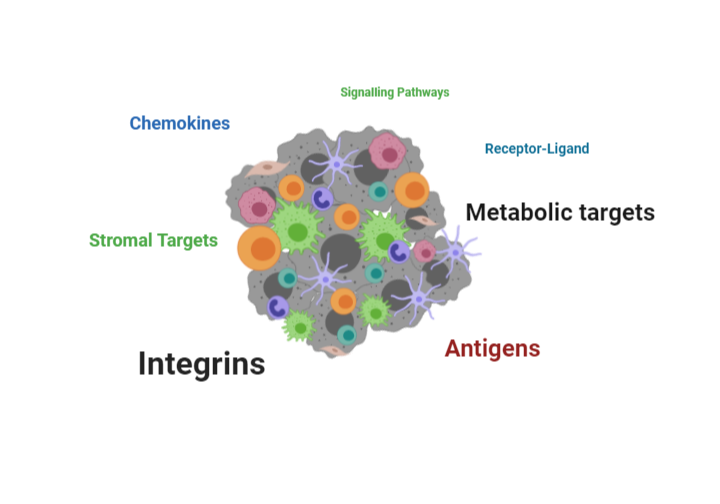
Screen based on environmental cues, cellular components and interactions

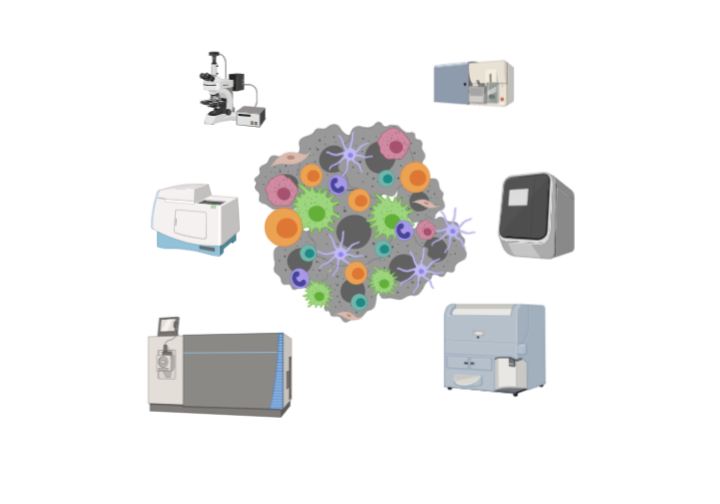
Leverage cutting edge instrumentation and equipment to analyze data
Our Process
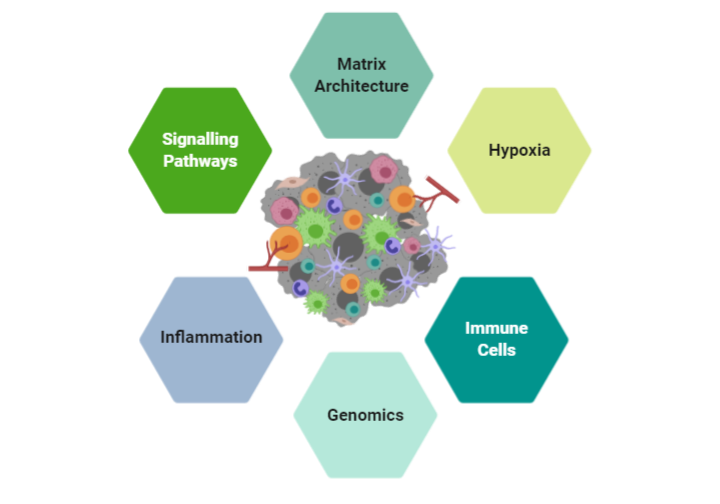
Screen Therapeutics we use a systematic process in identifying therapeutic targets and biomarkers.
- We utilize a set of proprietary HLA-matched, autologous (from the same patient) T cell and tumor cell lines. Cell lines are the foundation of any assay or drug discovery platform. These cell lines are well characterized with genomic and RNAseq data that serve as valuable “inputs” to interrogate the TME
- We utilize our cell lines in a proprietary ex vivo platform that is efficient and reproducible in capturing the patient’s TME and the extracellular matrix components that govern the motility and migration of immune cells into the tumor bed.
- Through collaborations with partners and academic institutions we leverage instrumentation, techniques and –omics platforms to validate our findings
- For novel targets – we confirm target receptors and ligands using human cell (transmembrane) arrays
- Confirm(and reproduce) our findings in in vivo models using our cell line platforms
- Expand findings into broad tumor types
- Pursue IND enabling studies to accelerate the target into clinical trials
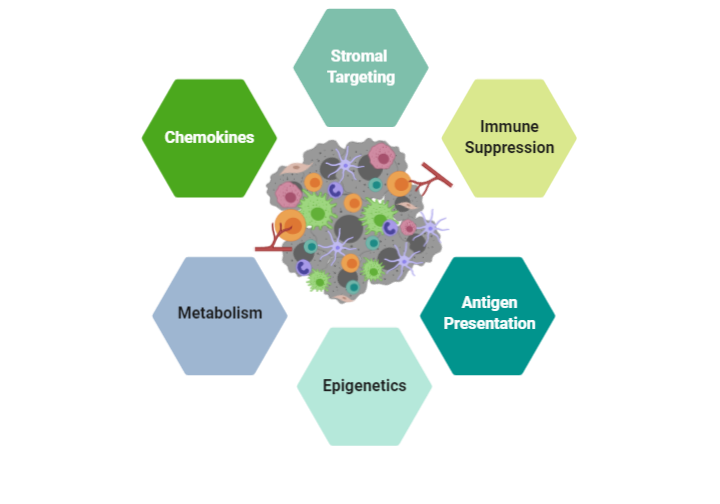
Our Pipeline
We have developed multiple executable research plans to accelerate the identification of novel targets and biomarkers.
Chemokines are heavily involved in the migration of lymphocytes and cells from early stages of development. Chemokines also have an important role in the development and maintenance of innate and adaptive immunity. Chemokines are involved in the pathobiology of chronic inflammation, tumorigenesis and metastasis, as well as autoimmune diseases. IMMUNE 3D® has established an extensive dataset of targeting chemokine and chemokine receptors and monitoring lymphocyte migration profiles in oncology using our proprietary cell lines. The chemokine profiles obtained in these studies can be applied across a broad spectrum of diseases.
Technology advancements and bioengineering of T cells have led to breakthrough therapeutics approaches such as Chimeric Antigen Receptor (CAR) T cells and T cell Receptor (TcR) that have demonstrated remarkable patient responses in oncology. The incredible success of these treatments have paved the way for further studies in other disease areas such as autoimmunity. The combination of our platform coupled with our genomic and TCR datasets we have obtained on our cell lines enables IMMUNE 3D® to identify novel or existing antigens on our T cells for further characterization and potential engineering into novel therapies.
Adhesion molecules play a major role in the recruitment of lymphocytes such as neutrophils to the site of inflammation. The activation and adhesion of lymphocytes through the endothelium is regulated by integrins and their ligands. The importance of adhesion molecules has been highlighted in rare diseases and recent studies have extended their role as potential targets in paving the way for lymphocytes to migrate through the extracellular matrix barriers. By using patient derived cells, IMMUNE 3D® can efficiently dissect and characterize the adhesion factors and integrins that may impede T cell or general lymphocyte migration as potential therapeutic targets for diseases.
Cancers often rapidly mutate and mutations in the genome can cause these malignant cells to express mutant proteins that are tumor specific and not expressed on normal cells, referred to as neoantigens. Thus, neoantigens are attractive immune targets because of their potential to elicit potent anti-tumor immune responses, albeit with lower potential for inducing autoimmunity. The 3D system can be used to uniquely expand tumor-specific T cell populations that respond to novel neoantigens. The 3D architecture provides a stringent environment for evaluating antigen presentation and in turn the migration and infiltration patterns of lymphocytes. Neoantigen presentation on tumors have been shown to play a role in the “hot” tumor environment and increase T cell infiltration. We have characterized our tumor cell lines for neoantigens and together with the HLA-matched T cell lines can observe migration patterns of lymphocytes that may yield novel receptor-ligand interactions and antigens for potential vaccine development.
Cancers often rapidly mutate and mutations in the genome can cause these malignant cells to express mutant proteins that are tumor specific and not expressed on normal cells, referred to as neoantigens. Thus, neoantigens are attractive immune targets because of their potential to elicit potent anti-tumor immune responses, albeit with lower potential for inducing autoimmunity. The 3D system can be used to uniquely expand tumor-specific T cell populations that respond to novel neoantigens. The 3D architecture provides a stringent environment for evaluating antigen presentation and in turn the migration and infiltration patterns of lymphocytes. Neoantigen presentation on tumors have been shown to play a role in the “hot” tumor environment and increase T cell infiltration. We have characterized our tumor cell lines for neoantigens and together with the HLA-matched T cell lines can observe migration patterns of lymphocytes that may yield novel receptor-ligand interactions and antigens for potential vaccine development.
Contact Us To Discuss Your Study
